The prize is awarded to the best students of the courses associated with the Department of Informatics of the Faculty of Sciences – University of Lisbon in a given year. Pedro Ruas received the prize for best student in the PhD Programme in Informatics in the year 2020/2021.
Author Archives: psruas@fc.ul.pt
LasigeBioTM – 2nd place at ProfNER Subtrack A
The LasigeBioTM team composed by Pedro Ruas, Vitor D. T. Andrade, and Francisco M. Couto achieved 2nd place at the Subtrack A of the ProfNER shared-task, which occurred in the context of the Social Media Mining for Health Applications (#SMM4H) ’21 Shared Task. Subtrack A was a tweet binary classification task, where the goal was to classify Spanish covid-19-related tweets according to the presence of entity mentions associated with professions, working statutes, and other work-related activities. The system developed by LasigeBioTM achieved an F1-score of 0.92, which was close to the result achieved by the top-performer system (0.93), and achieved the highest precision, 0.95. The complete results are available in the task overview paper.
The paper describing the developed system “Lasige-BioTM at ProfNER: BiLSTM-CRF and contextual Spanish embeddings for Named Entity Recognition and Tweet Binary Classification” is included in the Proceedings of the Sixth SMM4H Workshop and Shared Tasks, and it is available here.
Poster presentation at the 6th Social Media Mining for Health (#SMM4H) Workshop – NAACL 2021
Pedro Ruas will present the poster “Lasige-BioTM at ProfNER: BiLSTM-CRF and contextual Spanish embeddings for Named Entity Recognition and Tweet Binary Classification” (co-authors Vitor D. T. Andrade, Francisco M. Couto) at the 6th Social Media Mining for Health (#SMM4H) Workshop (June 10th, 17:30-18:15 GMT), co-located at the 2021 Annual Conference of the North American Chapter of the Association for Computational Linguistics (NAACL 2021).
Presentation in ECIR 2021 Doctoral Consortium: Deep Semantic Entity Linking
Presentation of the paper “Deep Semantic Entity Linking” in ECIR 2021 Doctoral Consortium by Pedro Ruas. Slides
Article: Linking chemical and disease entities to ontologies by integrating PageRank with extracted relations from literature
The paper “Linking chemical and disease entities to ontologies by integrating PageRank with extracted relations from literature”, co-authored by Pedro Ruas, Andre Lamurias and Francisco Couto was accepted for publication at Journal of Cheminformatics (h5-index of 41).
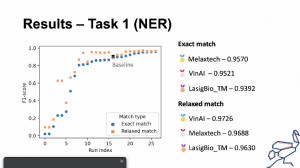
DeST participation results in Task 1 (NER) of ChEMU – 3rd position achieved
ChEMU consists of information extraction tasks over chemical reactions described in patents (https://chemu-patent-ie.github.io/). The goal of the task 1 (NER) was to identify chemical compounds and other entities involved in chemical reactions (like time, temperature, yield) in patents text and to assign a type to each entity according to its role in the context of the reaction.
The model developed by the DeST team (lasige_BioTM) outperformed the baseline model and was the third best result:
Exact evaluation
| Precision | Recall | F1-Score | |
| Baseline | 0.9071 | 0.9071 | 0.8893 |
| DeST | 0.9327 | 0.9457 | 0.9392 |
Relaxed evaluation
| Precision | Recall | F1-Score | |
| Baseline | 0.9208 | 0.9504 | 0.9354 |
| DeST | 0.9583 | 0.9960 | 0.9768 |